:focus(670x325:671x326))

UEA scientists are measuring some of the fastest processes on the planet to determine the effect that light has on proteins in living organisms, and the resulting structural changes that activate genetic switches.
In this important research project, Professor Steve Meech and his group at UEA with colleagues at the Central Laser Facility Harwell and Stony Brook University, New York, are studying the mechanism of operation of blue light sensing proteins. In their natural environment these proteins allow a wide variety of organisms to respond to light. For example, photosynthetic bacteria regulate their production of bacteriochlorophyll in response to the amount of light they are exposed to, or plants bend and move leaves to optimise their use of sunlight (phototropism). Currently little is understood about exactly how these proteins sense light and turn it into a signal to the host organism. This international research collaboration allows us to combine the very diverse fields of advanced laser spectroscopy, ultrafast chemical kinetics and chemical biology to address this biochemical problem.
By understanding this process, scientists can begin to place light sensitive proteins into organisms where they are not normally found, and exercise this ‘genetic switch’ as a useful tool – using tiny pulses of light to ‘switch’ a gene on or off and control how it reacts to its environment.
This ability to look at a very specific point in time and space within molecules, and measure such fast changes, could help us gain a better understanding of individual gene function and its reaction to light as part of a growing field of Optogenetics research.
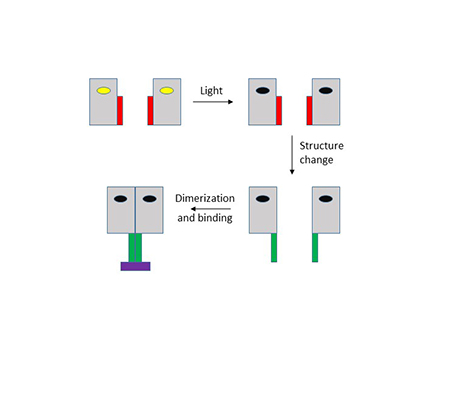
Professor Meech’s research group has been using pulses of light from lasers to direct photons at two classes of these proteins, BLUF and LOV domains. BLUF domains are found within about a quarter of all bacteria, while LOV domains are found even more widely, in bacteria, plants and fungi. In both cases, the protein domains bind a special blue light absorbing molecule, a flavin. Absorption of light by the bound flavin causes an immediate modification to the strength of the hydrogen bonds between it and its protein binding site. This initial step takes less than a pico second (one trillionth of a second) to occur. In BLUF domains this modified interaction initiates a wider structure change in the protein, occurring on the microsecond timescale. In LOV domains there is an extra step in which the photoexcited flavin undergoes a chemical reaction with a nearby amino acid residue (a cysteine) within a few microseconds. This reaction causes a shape change in the binding pocket which in turn gives rise to a structure change in less than a millisecond. These structure changes are sensed by other interacting proteins or nucleic acids, which causes the organism to respond accordingly; the process is illustrated in the diagram below.

Meech and co-workers are using ultrafast infrared (IR) spectroscopy to track the structural changes within the proteins. They use a pulse of blue light less than one ten trillionths of a second long to initiate the structure change, then they send a second equally short pulse of IR light through the sample to record its transmission spectrum. The delay between those two pulses can be adjusted between from one trillionth out to one thousandth of a second, allowing the time course of the structure change to be recorded through changes in the IR transmission spectrum. This provides the timescale for structure change. To provide insights into the mechanism the team make point mutations, modifying specific amino acids which are suspected of being involved in the structure change. The effect of these mutations on the timing and the spectrum associated with structure change are measured. In the protein many amino acids may be involved, so to provide further detail some natural amino acids are exchanged for unnatural amino acids. For example, an amino acid with a very prominent IR transition, will make their role easy to spot. Alternatively, a more subtle change is possible by picking an amino acid which has a change in its H-bond strengths, which then modifies the rate of the light induced structure change. These and similar modifications provide unprecedented detail on the mechanism of blue light sensing.
Understanding how these proteins help living organisms to sense their environment is interesting in its own right. However, in more recent years an important application for light sensing proteins has been found. Life scientists can now clone these light sensitive proteins and express then in a variety of organisms where they are not normally found. In this case the ‘genetic switch’ can be turned on by light activation in a way which is both temporally and spatially localised. This allows the researcher to use imaging methods to investigate the role of a specific gene, both where it acts and how long it takes to act, within a living cell. This branch of science, optogenetics, is in its infancy but is already providing exciting new data. The ability of this collaborative research program to understand and optimise the blue light response will support the growing area of optogenetics research.