Description
Lasso is a heuristic for computing edge-weighted rooted (super)trees from partial distance data under the assumption that the distances satisfy the molecular clock assumption [1]. It is based on theoretical work carried out in [2] and [3]. Rather than estimating missing distance values using some imputting scheme it exploits redundancy in the distance matrix to reconstruct a unique (in a well-defined sense) tree. The found tree can then be either displayed on a computer screen or saved in a .tex file which allows inclusion in a latex document (see the manual). Alternatively the found tree can also be output in Newick format which allows reading it by other phylogenetic tree reconstruction tools such as PHYLIP. Also and depending on the amount of missing information, the returned tree is not guaranteed to contain all taxa. See for [4] details.
If you use Lasso please cite [4] ie
Reconstructing (Super)trees from data sets with missing distances: Not all is lost, G. Kettleborough, J. Dicks, I. N. Roberts and Dr. Katharina T. Huber (2015) Mol. Biol. Evol. (doi:10.1093/molbev/msv027)
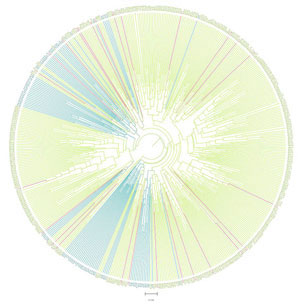
Fig 7 from [4] -- An equidistant supertree built by Lasso on two overlapping wheat data sets considered in [4]. For details concerning both data sets as well as the colour coding see [4].
Manual
A basic manual for lasso is available
Availability
Download Lasso as a zip file, containing the binaries, read me and example file
Test files
The yeast data set considered in [4].
Support
For questions please contact Katharina Huber
References
[1] Molecular disease, evolution, and genic heterogeneity, E. Zuckerkandl, L.B. Pauling, In:
Kasha M, Pullman B, editors. Horizons in biochemistry.New York: Academic Press. (1962) pp 189–225.
[2] Lassoing and corralling rooted phylogenetic trees, K. T. Huber, A-A Popescu, Bulletin of Mathematical Biology
2013, 75(3), pp 444-465
[3] Distinguished minimal topological lassos, K.T. Huber, G. Kettleborough. Siam Journal on Discrete Mathematics (in press).
[4] Reconstructing (super)trees from data sets with missing distances: not all is lost, G. Kettleborough, J. Dicks, I.N. Roberts, Dr. K.atharina T. Huber (2015) Mol. Biol. Evol. (doi:10.1093/molbev/msv027)

)