MIfold: Predicting RNA structure using mutual information
Overview
MIfold is a matlab toolbox that uses mutual information and related measures to infer and display secondary structures (including pseudoknots). Given a sequence alignment MIfold computes and displays the mutual and sequence information of the alignment. MIfold also uses a dynamic programming algorithm to predict the secondary structure with maximal total mutual information.
The algorithm is implemented in Matlab6.5 and it accompanies our Applied Bioinformatics paper;
Freyhult, Eva, Moulton, Vincent and Gardner, Paul, 2005 Predicting RNA structure using mutual information, Applied Bioinformatics. 4(1):53-9.
Download and installation
MIfold has been tested under Matlab 5.3 and Matlab 6.5 both on unix and windows.
The MIfold package can be downloaded as a zip file.
The distribution contains the directory mfiles, a tutorial, a README file and an example alignment. The directory mfiles contain all the matlab source files, the README file contains instructions of how to install and run MIfold both for standard usage and more advanced use. In the README file all the .m-files in the distribution is shortly described. For more information of any of the MIfold functions type help filename in the matlab command window.
To run MIfold
Unpack the distribution and open matlab. Add the search path to the MIfold .m-files by typing the following line in the matlab command window:
>> addpath /home/user/MIfold/mfiles
For simplicity (if you want to use MIfold more than once) you should add this line to your startup.m file.
To launch the MIfold GUI just type MIfold in the matlab command window.
>> MIfold
For instructions on how to use MIfold please read the README file and the tutorial (PDF, 82KB) that accompanies the distribution. For questions about the installation, bug reports etc. please contact Eva Freyhult.
Screenshots
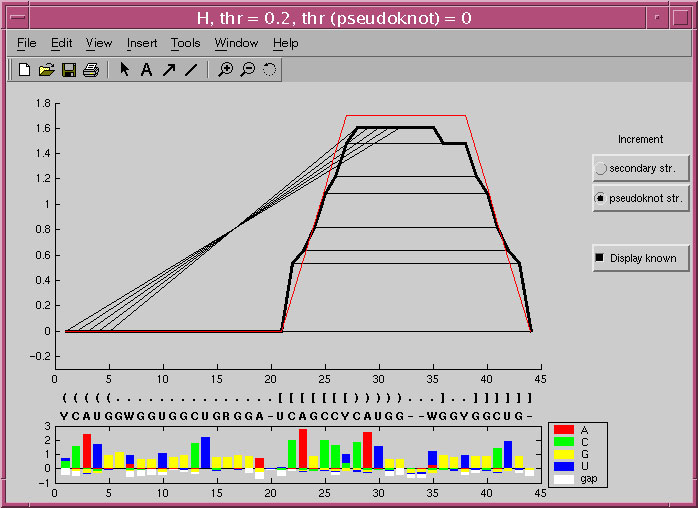
A screenshot of the MIfold display of the prediction of the PRP pseudoknot structure.
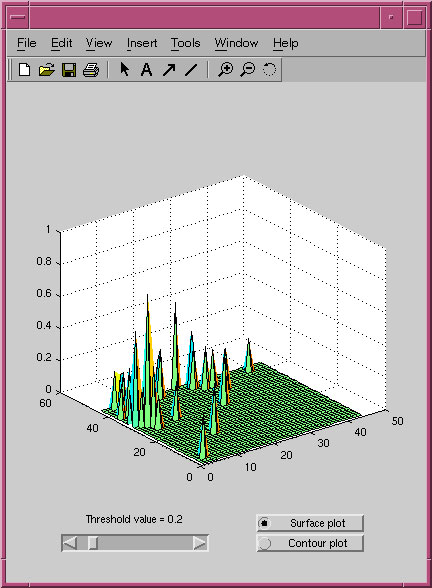
A screenshot of the display of the mutual information distribution for the PRP pseudoknot alignment.
Disclaimer
This software is supplied as-is, with no warranty of any kind.
Research Team

)