Introduction
TriLoNet (Trinet Level One Network) is a Java software package created to construct rooted level-1 phylogenetic networks from aligned DNA sequence data using a trinet-based approach as described in [1]. It contains the following main module:
TriLoNet - constructs level-1 phylogenetic networks from a multiple sequence alignment, or a dense collection of trinets.
In addition, it also contains the following four complementary tools including:
SeqTrinet - constructs a dense set of trinets from a multiple sequence alignment
TriExtract – extracts the set of trinets from a level-1 phylogenetic network
PNDist – calculates the trinet and the Robinson-Fould distances between two level-1 phylogenetic networks
PNGenerator – randomly generates a set of n level-1 phylogenetic networks on l leaves
The algorithms and details for the last three tools can be found in [2].
Citation
If you use this software, please cite either [1] for network construction, or [2] for complementary tools
Basic Usage
To get started, extract the contents of TriLoNet.zip and navigate to the location of the TriLoNet.jar file using the command prompt/terminal. To run TriLoNet from the command line, use
java -jar TriLoNet.jar inputFile
where the inputFile could be a multiple DNA sequence alignment (in nexus format or fasta forma), or a dense trinet (.TNETS files). The result is contained in output.dot (open in GraphViz) and output.txt which contains the eNewick string of the constructed phylogenetic network as well as a summary of the construction process. The file output.tree will contain the eNewick string representation of the network constructed by TriLoNet. Additionally, the names of the output files can be specified when running TriLoNet, for example:
java -jar TriLoNet.jar example.nex anExample.dot exampleOutput.txt
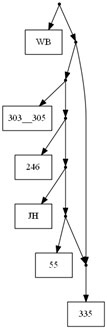
Example
The website phylonetworks hosts several data sets containing recombinants. Here we have included an example on a giardia data set studied in Cooper et al.(2007). In this data set the taxon 335 is a recombinant. The line
java -jar TriLoNet.jar Cooper.nex --b5979,7444
will result in the network shown below.
Complementary Tools
 SeqTrinet will read in a multiple sequence alignment and output a dense collection of trinets. To use the SeqTrinet module, use the argument --st
SeqTrinet will read in a multiple sequence alignment and output a dense collection of trinets. To use the SeqTrinet module, use the argument --st
java -jar TriLoNet.jar inputFile --st
where the inputFile contains a multiple DNA sequence alignment (in nexus format .nex or fasta forma .fsa).
TriExtract will extract the trinets displayed by a level-1 phylogenetic network. To do this the user can specify an input .tree file containing an eNewick string and an output .tnets file to write to and record the set of trinets. Entering
java -jar TriExtract.jar firstExample.tree
into the terminal will instruct TriExtract to construct the network represented by the eNewick string contained in firstExample.tree and output the set of trinets displayed by this network.
PNDist calculates the trinet distance and the Robinson-Fould distances between a pair of binary rooted level-1 phylogenetic networks (see [2] for details). PNDist accepts as input a .tree file containing two eNewick strings on separate lines. As an example, entering
java -jar PNDist.jar example.tree
will produce a text file containing scores of D'tn = 0.6202 (for the normalized trinet distance) and D'rf = 0.3448 (for the normalized Robinson-Foulds distance).
PNGenerator randomly generates rooted binary level-1 phylogenetic networks and takes in two integers as input. One of the input arguments l specifies the number of leaves wanted in the generated network. The other input argument n specifies the number of random networks created by PNGenerator. As an example, entering
java -jar PNGenerator.jar --l50 --n100
into the terminal will instruct PNGenerator to generate 100 random networks each on 50 leaves.
Availability
Trilonet together with complementary tools can be downloaded from here Trilonet.zip. Detailed installation and usage instructions can be found in the manual, which is distributed together with the program and can also be downloaded here Trilonet_manual.pdf.
Test files illustrate the different types of input data for Trilonet and the complementary tools are available for download together with the program. The three datasets, Lev(1), Lev(10), and Ran mentioned in [2] are available here.
Disclaimer
This software is supplied as-is, with no warranty of any kind expressed or implied. We have made every effort to avoid errors in design and execution of this software, but we will not be liable for its use or misuse. The user is solely responsible for the validity and consequences of any results generated. This program is distributed in the hope that it will be useful, but without any warranty; without even the implied warranty of merchantability or fitness for a particular purpose. See the GNU General Public License for more details.
References
- Oldman, J., Wu, T., van Iersel, L., and Moulton, V., TriLoNet: Piecing together small networks to reconstruct reticulate evolutionary histories. Molecular Biology and Evolution, in press.
- Moulton V., Oldman, J., and Wu, T., Comparing networks by counting trinets. Preprint.
Research Team
James Oldman, Prof. Vincent Moulton and Dr. Taoyang Wu
For technical questions, please e-mail james.oldman@gmail.com
TriLoNet, TriExtract, PNDist and PNGenerator are Copyright 2011-2016 James Oldman, Vincent Moulton and Taoyang Wu.

)