Background
Reticulate evolutionary signal can arise from processes that act on both the species and population level. An instance of the latter process is introgression, which provides an important source of innovation for evolution. Many approaches aimed at representing introgression rely on a phylogenetic-network-based model that was originally developed for lateral gene transfer between species. However, this can be problematic since the constraints that govern introgression are fundamentally different from those governing lateral gene transfer. Exploiting the fact that in many cases lineage information is available and that it is sometimes identifiable in terms of alleles of a gene, we propose to represent introgression in terms of a certain "overlay" of the lineages and an allele tree. Such an overlay or Overlaid Species Forest (OSF for short) is a multiply rooted phylogenetic network. Here, we present a novel algorithm for building such networks.
Description
OSF-Builder is an implementation, in Python, of the OSF-Builder algorithm. It runs on python2, and requires the packages Given a species forest, that is, a set F of lineage trees on some set of species, and an allele tree for these species, the algorithm constructs an "overlay" of the latter into the forest, called an Overlaid Species Forest (or OSF for short), that minimizes the number of arcs across distinct trees. For more information about the OSF-Builder algorithm and its properties, we refer to:
- A.-A. Popescu, G. E. Scholz, M. I. Taylor, V. Moulton and K. T. Huber. OSF-Builder: A new tool for reconstructing and representing phylogenetic histories involving introgression.
A preprint of the paper may also be obtained by directly contacting the corresponding author, Dr. Katharina T. Huber.
Directions of use
Download the program into a folder which contains the following input files:
The species forest: n>0 species trees in newick format, named from l1.tre to ln.tre. The order matters, so the user can test different orderings to compare outputs. For more consistent results, we suggest to sort them by age, the oldest lineage coming first and the most recent one coming last, if such information is known.
The allele tree: A tree in newick format, named gene.tree.
The allele-species map: A text file called gene_map.txt in which each line consists of two labels: the first one being a leaf in the gene tree (all must appear exactly once), the second one a leaf in the forest.
Outputs
Two files are generated, both representing the OSF induced by the input.
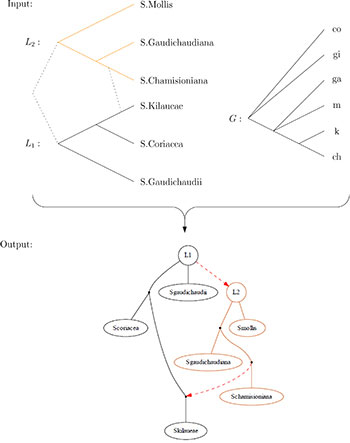
- The first file is called osf.tree. It is a network in eNewick format, which can be read using popular softwares such as Dendroscope (Huson et. al.) or SplitsTree. It consists of the trees l1 to ln, whose roots have been artificially joined up with a common new root for technical reason, to which contact arcs, representing potential introgression events, have been added.
- The second file is called osf2.gve, and can be read using e. g. the GraphViz softwares. It provides a graphical representation of a constructed OSF, in which all trees of the forest have different colors, and the contact arcs are represented as red, dashed arrows. The GraphViz software is then able to export that graphical representation as e. g. a .pdf or a .jpeg file.
Availability
OSF-Builder is freely available and may be downloaded as a zip file which contains:
The Python source code, OSF-Builder.py.
A folder synth containing a synthetic example, in the form of two species trees l1.tre, l2.tre and an allele tree gene.tree, all three in newick format, and a text file gene_map.txt.
A folder Heliconious, containing the biological data set used in the associated paper. It contains seven species trees, named from l1.tre to l7.tre in newick format, an allele tree gene.tree in newick format, and a text file gene_map.txt.
Research Team
Dr. A.-A. Popescu, G. Scholz, Dr. M. Taylor, Prof. Vincent Moulton, Dr. Katharina T. Huber

)