Taylor lab - Research
Current and recent research projects
Evolutionary biology of Neotropical catfishes

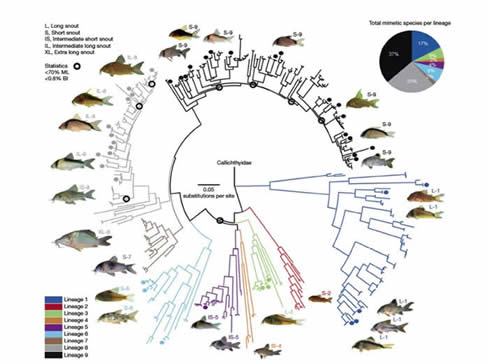
The Corydoras are a fascinating group of South American catfishes. They are the most species rich genus of all catfishes comprising some 150 described species with many further species either undescribed or awaiting discovery. Alongside the taxonomic richness, the group displays enormous karyotypic diversity and genome size variation with chromosomal counts ranging from 2n=34 to 2n=134, and haploid C values ranging from 0.75pg to 4.4pg. I am interested in a range of questions in this group that includes the evolution of colour patterns and mimicry, coexistence, the evolution of genome size variation, patterns of diversification rate variation across the group.

Key papers: Alexandrou, M. A., C. Oliveira, M. Maillard, R. A. R. McGill, J. Newton, S. Creer and M. I. Taylor. Competition and phylogeny determine community structure in Müllerian co-mimics. Nature, 469, 84–88, 2011. DOI:10.1038/nature09660
See also Nature News and Views in same issue.
Collaborators: Markos Alexandrou, Claudio Oliveira ((UNESP, Brazil); Ian Fuller (Catfish Study Group, UK).
Funding: NERC small grant (NE/C001168/1), NERC Mass Spectrometry access grant (NE/F007205/1) (Taylor, MI); NERC PhD studentship (NE/F007205/1) (M. Alexandrou).
Fisheries Induced Evolution

Understanding the drivers behind changes in individual size and age of maturation in exploited fish stocks is an important step to long-term fisheries management. However, despite the increasing incidence of such shifts, the relative contributions of genetic and environmental factors remain uncertain and are much-debated. Using the Trinidadian guppy (Poecilia reticulata) as a model system to examine the genetic basis of fisheries-induced evolution, recently graduated PhD student Serinde van Wijk identified significant genetic changes associated with changes in life histrory traits. The work is now being extended by Hazel Perry who will examine in more detail the genomic shifts associated with artificial selection in the guppy system.
Key papers: van Wijk, SJ, Taylor, MI, Creer, S, Dreyer, C, Rodrigues, FM, Ramnarine, IW, van Oosterhout, C and Carvalho, GR (2013) Experimental harvesting of fish populations drives genetically-based shifts in body size and maturation. Frontiers Ecol. Environ. Online early e-view http://www.esajournals.org/doi/abs/10.1890/120229

AquaTrace - The development of tools for tracing and evaluating the genetic impact of fish from aquaculture: "AquaTrace"
Project Description: AquaTrace is an FP7 funded project running from 2012 - 2016. Aquaculture represents an important solution to meet the escalating demand for fish. The development of appropriate legislation within the European Union aquaculture sector underpinned by cutting‐edge research and technology. This necessitates implementation of breeding programmes and farming technologies which are economically viable, environmentally friendly, and perceived as socially acceptable. The rationale behind AquaTrace is development of reliable and cost‐effective molecular tools to identify of the genetic origin of both wild and farmed fish (assignment and genetic traceability), as well as for the detection of interbreeding genetic introgression between farmed and wild stocks. This work will be carried out
on three marine fish of economic significance: the European sea bass (Dicentrarchus labrax), gilthead sea bream (Sparus aurata), and turbot (Scophthalmus maximus). To address quantitative effects of farm introgression, the rationale is to examine links between key fitness and life‐history traits and
specific functional genetic variation between wild and farmed fish, using Atlantic salmon and brown trout as model species. Thus, the scientific objectives of AquaTrace are to address and assess the genetic impact of aquaculture escapees introducing genes to wild populations that have been undergoing adaptation to farmed conditions through breeding and domestication selection.
See project website here
Celtic Sea Trout project (CSTP)(website)

The Celtic Sea Trout Project is a groundbreaking, €2 million, multi-agency partnership investigation into the sea trout stocks and fisheries of the rivers entering the Irish Sea. It is part-funded by the EU INTERREG V1A Ireland-Wales Programme with additional support from government agencies, voluntary bodies and private fishery interests in Wales, Ireland Southwest Scotland, Northwest England and the Isle of Man. The active fieldwork programme will take place over three years from April 2010.
The project is led by Bangor University with the other main partner the Inland Fisheries Ireland. Other partners include: Association of Rivers Trusts, Solway Sea Trout Group, Clwyd and Conwy Fisheries Trust, Carmarthenshire rivers Trust, Teifi Rivers Trust, Slaney Rivers Trust, Loughs Agency, AFBI, University College Cork, Queens University Belfast, Marine Institute, Countryside Council for Wales, Department for Culture Arts and Leisure (NI).
The Celtic Sea Trout Project aims to:
To understand and describe sea trout stocks in the Irish Sea and thereby to enhance sea trout fisheries and strengthen their contributions to quality of life, to rural economies and to national biodiversity.
To explore the use of sea trout life history variation as a tool to detect and understand the effects of climate change.
The MEFGL is involved with the genetics and life history components of the project using microsatellite and SNP markers to investigate genetic structure among sea trout populations on the Irish Sea side of the UK.
Contacts: Niklas Tysklind (postdoctoral researcher)(bssc08@bangor.ac.uk)

Population structure of cod around the UK: scale, mechanics and dynamics
![]() PIs: Prof. Gary Carvalho and Dr. Martin I. Taylor, in collaboration with Prof. William Gurney and Dr. Douglas Speirs (University of Strathclyde), Dr. Clive Fox (Scottish Association of Marine Science).
PIs: Prof. Gary Carvalho and Dr. Martin I. Taylor, in collaboration with Prof. William Gurney and Dr. Douglas Speirs (University of Strathclyde), Dr. Clive Fox (Scottish Association of Marine Science).
This NERC funded project aims to investigate population structure of cod in UK waters using single nucleotide polymorphism markers (SNPs), Otolith microchemistry and mathematical modeling. The project includes three major UK fisheries agencies (Marine Scotland, CEFAS, AFBI) with partners in UK Universities, a NERC Institute and a Danish partner (DTU-Aqua).
Population structure and traceability of marine fishes (FishPopTrace).

FishPopTrace is an international project aiming at the construction of a Pan-European framework, built on advanced technologies, for product traceability and policy related monitoring, control and surveillance (MCS) in the fisheries sector. Pursuing a holistic approach, FishPopTrace can contribute to fisheries management and conservation measures in line with the global attempt to move towards sustainable fisheries.
The project was coordinated from Bangor.
An additional small project developing SNP markers for hake using Solexa sequencing of cDNA complements FishPopTrace: MerSnip – Development of SNP markers for hake. Funded by the Joint Research Council (JRC). Collaborators include Rob Ogden (Trace Network), Frank Panitz (Arhus, Denmark).
 FishPopTrace booklet available here
FishPopTrace booklet available here
Aquagen - Genetics for Marine Aquaculture Management (website)
The genetic assignment of fish to their farm of origin is key to resolving fundamental questions and challenges arising from aquaculture. It constitutes an asset to future traceability and (eco-) labelling schemes, as proposed by the Commission and the European Parliament. Moreover tracing back aquaculture escapees to the farm of origin is indispensible to evaluate the potential genetic impact of domestication of farmed aquatic species on wild populations, as well as for control purposes. Fish farming can also be used to release cultured juveniles for restocking of wild populations and sea ranching. Assessing the success of such approaches also requires the ability to trace farm origin individuals back to the source.
By focussing on two marine fish species, of major commercial interest, common sole (Solea solea) and Atlantic cod (Gadus morhua), this study aims at building a basis for aquaculture management of marine fish based on genetic tools, including the monitoring and control of farm escapees, and fish farming for restocking purposes of wild populations.
Aquagen will:
1. Evaluate the feasibility of distinguishing genetically between farmed and wild common sole and Atlantic cod;
2. Evaluate the feasibility of genetically tracing individual fish back to a farmed origin, - ideally to the farm of origin -, in common sole and Atlantic cod;
3. Develop appropriate identification methodologies, including a comparison of population assignment methods with parentage identification techniques, to provide the greatest power for correctly identifying farmed individuals;
4. Develop traceability assays within a forensic framework (standard operating procedures) in order to ensure that the assays can be applied in a legal context;
5. Examine the applicability of the approach to other marine aquaculture species, particularly farmed in the Mediterranean, e.g. European seabass (Dicentrarchus labrax) and gilthead seabream (Sparus aurata).
The outcomes are expected to support the enhancement of marine aquaculture under the Common Fisheries Policy remit.


Identification and mapping of planktonic fish eggs
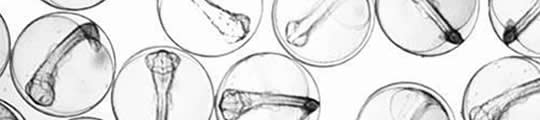
Identifying the early stage eggs of a number of commercially important fish species occuring in the waters surrounding the UK is impossible prior to the development of larval pigmentation. In collaboration with Dr Clive Fox at the CEFAS Laboratory in Lowesoft, and with Defra and EU funding, the development of a genetic assay1 to distinguish between early stage cod (Gadus morhua), haddock (Melanogrammus aeglefinus L.) and whiting (Merlangius merlangius) eggs has facilitated the accurate mapping of spawning grounds of these species in the Irish2 and North Seas3. The assay will also allow the future application of catch independent stock asessment methods such as the Annual Egg Production Method (AEPM).
Key publications: Fox, CJ, Taylor, MI et al. (2008) Mapping the spawning grounds North Sea cod (Gadus morhua) by direct and indirect means. Proceedings Royal Society London B. 275: 1543-1548.
Fox, C.*, Taylor, M. I.*, Pereyra, R., & Rico, C. (2005) Mapping of the spawning grounds of Irish Sea gadoids using genetic identification of planktonic eggs. Molecular Ecology 14, 879-884. * Joint first authors.
Molecular detection of predator prey relationships “PREDATE”
During their egg and larval stages, marine fish experience high levels of mortality, with mortality rates for temperate species typically between 5-20% per day. Since the duration of the egg larval and juvenile phases extends over several months, this leads to cumulative mortalities of 98-99%. A consequence of these high mortality rates is that small shifts in daily mortality rate accumulate to generate large changes in the numbers of survivors. This project focuses on one aspect of egg and larval mortality – predation. In this project we are developing molecular tools (Taqman probes) for detecting the presence of eggs and larvae of several commercial species in the stomachs of predators including fish (e.g. herring and sprat), crustacea (e.g. mysids) and gelatinous predators (e.g. the moon-jellyfish - Aurelia). Several of these predators have undergone recent large changes in abundance, for example, since the 1980s the herring stock in the North Sea has undergone a significant recovery and several studies suggest that the abundance of gelatinous predators may also be increasing. Such changes may be linked to recent warming of the North Sea and changes in its zooplankton composition. These changes have the potential to exert increased mortality on early life history stages of commercial fish such as cod and plaice and damage stock viability or inhibit stock recovery.
Funding: Defra (MF0156) “PREDATE”; Fisheries Society of the British Isles (small grant): Detecting predation on flatfish nursery grounds using modern molecular techniques PI: Dr. Aitor Albaina Vivanco and Dr. Martin Taylor in collaboration with Dr. Clive Fox, Scottish Association for Marine Science Funded by The Basque Government
Key publications:
Fox C.J., Taylor, M.I., van der Kooij, J., Taylor, N., Milligan, S.P., Albaina, A., Pascoal, S., Lallias, D., Maillard, M., Hunter, E. (2012) Identification of marine fish egg predators using molecular probes. Marine Ecology Progress Series. 462:205-218 - doi:10.3354/meps09748.
Hunter, E., Taylor, N., Fox, C. J. Maillard, M. & Taylor, M. I. (2012) Effectiveness of TaqMan probes for detection of fish eggs and larvae in the stomach contents of a teleost predator. Journal of Fish Biology 81(1):320-8. doi: 10.1111/j.1095-8649.2012.03298.x
Albaina, A; Fox, CJ; Taylor, N; Hunter, E; Maillard, M and Taylor, MI. (2010) A TaqMan real-time PCR based assay targeting plaice (Pleuronectes platessa L.) DNA to detect predation by the brown shrimp (Crangon crangon L.) and the shore crab (Carcinus maenas L.) Assay development and validation. Journal of Experimental Marine Biology And Ecology, 391: 178-189.
Albaina, A., Taylor, M. I., Fox, C. Predation impact on recently settled juvenile plaice (Pleuronectes platessa) estimated by visual and molecular means. Marine Ecology Progress Series. In press.
Current PhD and Masters projects
Evolution of Corydoradinae catfishes
NERC funded PhD project (2011-2014)
Corydoras are neotropical catfishes that are particularly interesting for evolutionary biologists. They are the most species rich genus of all catfishes with more than 170 described species, demonstrate multiple genome duplication events across the phylogeny, with genome duplication and subsequent diversification rate correlated, and also exhibit complex mimetic colour pattern relationships both with congeneric taxa and outside the group. This project will use a multidisciplinary approach to investigate factors that may increase speciation rates (or decrease extinction rates) in polyploid taxa compared to diploids.
Contact: Sarah Marburger (bspe08@bangor.ac.uk)
Fisheries induced evolution
A PhD project (2012-2014) is examining the genetic basis of fisheries-induced evolution, using the Trinidadian guppy, Poecilia reticulata, as a model system. Selection experiments aimed at replicating the effects of size selective mortality are being conducted, and the underlying genetic response at candidate loci monitored. Contact: Hazel Perry (hazelperry@hotmail.com)
Evolution of African cichlid fishes
Alex Tyers (bspa36@bangor.ac.uk)
Alex is investigating mate choice and hybridization in Lake Malawi cichlids. I am co-supervising Alex with Professor George Turner.